Last weekend, I had the opportunity to attend a fascinating workshop on generative methods in drug discovery. The workshop was led by two experts in the field - Stanislaw Jastrzebski, CTO and Chief Scientist at Molecule.one, and Tomasz Danel, lead machine learning scientist at Insitro.
It was run as part of the ML in PL conference
Overview of the Workshop
The day started with an overview of the traditional drug discovery process and how ML can accelerate certain steps. The presenters talked about:
- Virtual screening using molecular fingerprints and QSAR modeling
- Retrosynthesis prediction to figure out how to synthesize molecules
- Molecular generation with deep learning
We then jumped into hands-on tutorials in Google Colab notebooks. We practiced basic workflows like:
- Sampling random compounds from ZINC (a database of readily purchasable compounds that can be used for virtual screening.) and testing their activity against a target protein
- Mutating top compounds by tweaking their SMILES strings
- Building QSAR models to predict activity using Scikit-Learn and PyTorch
Next we leveled up to more advanced ML techniques:
- Implementing an active learning loop that iteratively collects data, retrains models, and proposes new compounds. This was super cool to see in action.
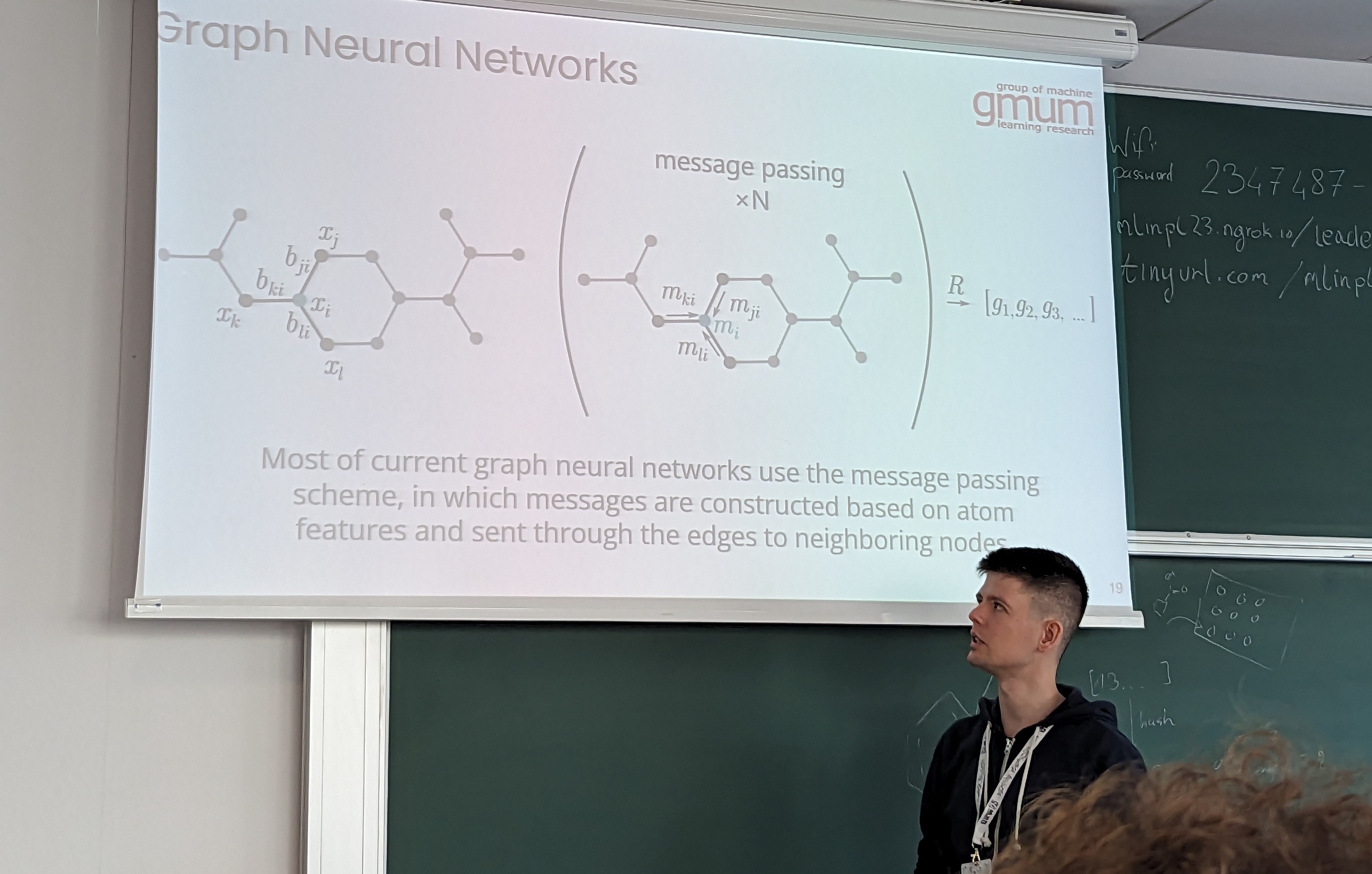
- Using graph neural networks on molecular graphs to build predictive models. I’m excited to explore GNNs more.
The workshop finished up with a friendly competition to find high-affinity binders for two drug targets. It was a race against the clock to come up with the best compounds.
Key Takeaways
Here are some of my biggest takeaways from the workshop:
- The end-to-end process of hit finding, lead generation, and optimization
- How to intelligently search chemical space by iterating on active compounds
- The power of combining machine learning with wet lab experiments
- How far deep learning has come in modeling molecular properties and activities
And we got to work with awesome Python libraries:
- RDKit for cheminformatics features
- Selfies for representing and mutating molecular graphs
- Scikit-learn for QSAR modeling
- PyTorch for graph neural networks
The instructors were phenomenal and I really appreciated the hands-on, practical nature of the content. I’m amazed at the high quality of the workshop and am immensely grateful to Stanislaw and Tomasz for their time and effort putting together an awesome workshop!